We will demonstrate distributed deep learning for the problem of anomaly detection.
The first step is to set up Elephas on Ubuntu OS.
The dependencies for Elephas are present in a file "req.txt" at the 'current working directory':
Flask==1.0.2
hyperas==0.4
pyspark==2.4.0
six==1.11.0
tensorflow==1.15.2
pydl4j>=0.1.3
keras==2.2.5
Next, we run following commands in Anaconda terminal:
conda create -n elephas python=3.7
conda activate elephas
pip install -r req.txt
pip install elephas ipykernal jupyter jupyterlab pandas matplotlib seaborn scikit-learn autoflake
python -m ipykernel install --user --name elephas
Check the elephas version:
(elephas) ashish@ashish-VirtualBox:~/Desktop$ pip show elephas
Name: elephas
Version: 0.4.3
Summary: Deep learning on Spark with Keras
Home-page: http://github.com/maxpumperla/elephas
Author: Max Pumperla
Author-email: max.pumperla@googlemail.com
License: MIT
Location: /home/ashish/anaconda3/envs/elephas/lib/python3.7/site-packages
Requires: hyperas, keras, cython, six, pyspark, tensorflow, flask
Required-by:
Python code from Jupyter Notebook:
from collections import Counter
import pandas as pd
import numpy as np
from pyspark import SparkContext
from elephas.spark_model import SparkModel
from elephas.utils.rdd_utils import to_simple_rdd
import keras
print("keras", keras.__version__)
import pyspark
print("pyspark", pyspark.__version__)
import tensorflow
print("tensorflow", tensorflow.__version__)
keras 2.2.5
pyspark 2.4.0
tensorflow 1.15.2
sc = SparkContext.getOrCreate()
df_iris = pd.read_csv('files_1/iris_flower/iris.data',
names = ['sepal_length', 'sepal_width', 'petal_length', 'petal_width', 'flower_class'])
for i in ['Iris-setosa', 'Iris-versicolor', 'Iris-virginica']:
print(i, ":")
print(df_iris[df_iris.flower_class == i].describe())
Iris-setosa :
sepal_length sepal_width petal_length petal_width
mean 5.00600 3.418000 1.464000 0.24400
min 4.30000 2.300000 1.000000 0.10000
max 5.80000 4.400000 1.900000 0.60000
Iris-versicolor :
sepal_length sepal_width petal_length petal_width
mean 5.936000 2.770000 4.260000 1.326000
min 4.900000 2.000000 3.000000 1.000000
max 7.000000 3.400000 5.100000 1.800000
Iris-virginica :
sepal_length sepal_width petal_length petal_width
mean 6.58800 2.974000 5.552000 2.02600
min 4.90000 2.200000 4.500000 1.40000
max 7.90000 3.800000 6.900000 2.50000
We will use the "max" values to introduce anomalies in the dataset.
df_iris_anomalies = pd.DataFrame({
"sepal_length": [7, 8, 9],
"sepal_width": [7, 4, 5],
"petal_length": [4, 6, 8],
"petal_width": [4, 3, 3],
"flower_class": ['Iris-setosa', 'Iris-versicolor', 'Iris-virginica']
})
df_iris = pd.concat([df_iris, df_iris_anomalies], axis = 0)
def frequency_encoder(input_df, column_name):
counter = 0
ranked_dict = {}
def ranker():
nonlocal counter
counter += 1
return counter
for i in Counter(input_df[column_name]).most_common():
ranked_dict[i[0]] = ranker()
return ranked_dict
ranked_dict = frequency_encoder(df_iris, column_name = 'flower_class')
df_iris['flower_class_enc'] = df_iris['flower_class'].apply(lambda x: ranked_dict[x])
X_train = df_iris[['sepal_length', 'sepal_width', 'petal_length', 'petal_width', 'flower_class_enc']]
Defining Model
act_func = 'tanh'
model = keras.Sequential()
model.add(keras.layers.Dense(5, activation=act_func, kernel_initializer='glorot_uniform',
input_shape=(X_train.shape[1],)))
model.add(keras.layers.Dense(4, activation=act_func, kernel_initializer='glorot_uniform'))
model.add(keras.layers.Dense(5, activation=act_func, kernel_initializer='glorot_uniform'))
model.add(keras.layers.Dense(X_train.shape[1], kernel_initializer='glorot_uniform'))
opt = keras.optimizers.Adam(lr=0.01)
model.compile(loss='mse', optimizer=opt, metrics=['accuracy'])
model.summary()
rdd = to_simple_rdd(sc, X_train, X_train)
spark_model = SparkModel(model, frequency='epoch', mode='asynchronous')
spark_model.fit(rdd, epochs=20, batch_size=32, verbose=0, validation_split=0.1)
Prediction
X_pred = spark_model.predict(np.array(X_train))
X_pred = pd.DataFrame(X_pred, columns=X_train.columns)
X_pred.index = X_train.index
scored = pd.DataFrame(index=X_train.index)
scored['Loss_mae'] = np.mean(np.abs(X_pred-X_train), axis = 1)
fraction_of_anomalies = 0.03
scores_threshold = scored['Loss_mae'].quantile([1 - fraction_of_anomalies])[1 - fraction_of_anomalies]
is_anomaly = scored['Loss_mae'] > scores_threshold
df_anomaly = X_train[is_anomaly]
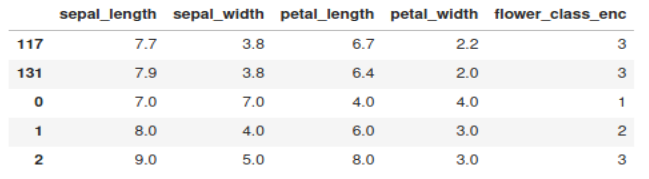
print(df_anomaly)
The three anomalies that we introduced in the dataset are appearing at the bottom.
Elephas Documentation: Github
Pages
- Index of Lessons in Technology
- Index of Book Summaries
- Index of Book Lists And Downloads
- Index For Job Interviews Preparation
- Index of "Algorithms: Design and Analysis"
- Python Course (Index)
- Data Analytics Course (Index)
- Index of Machine Learning
- Postings Index
- Index of BITS WILP Exam Papers and Content
- Lessons in Investing
- Index of Math Lessons
- Index of Management Lessons
- Book Requests
- Index of English Lessons
- Index of Medicines
- Index of Quizzes (Educational)
Friday, July 31, 2020
Distributed Deep Learning Using Python Packages Elephas, Keras, Tensorflow and PySpark
Subscribe to:
Post Comments (Atom)
No comments:
Post a Comment